For further inquiries and other sample submission requirements for Next Generation Sequencing (Ion Torrent PGM, Ion Torrent Proton, or Illumina MiSeq), kindly contact us or send us an e-mail to dnasequencing@pgc.up.edu.ph.
For inquiries regarding NextSeq 500, please contact us through pcari.sgcl@gmail.com. We'll be glad to assist you!
Whole Genome Sequencing Sample Requirements
Illumina MiSeq
-
Submit at least 80 uL of 50 ng/uL gDNA in water (concentration should be based on QuBit Fluorometer).
-
A260/A280 ratio of DNA samples should be at ~1.8-2.0.
-
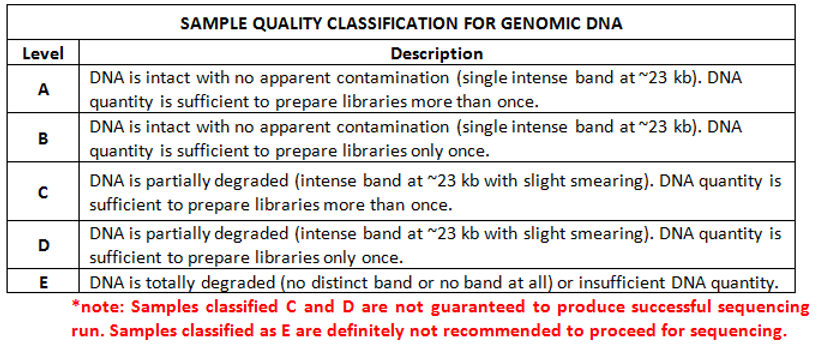
Please refer to the agarose gel image below for sample quality guideline.

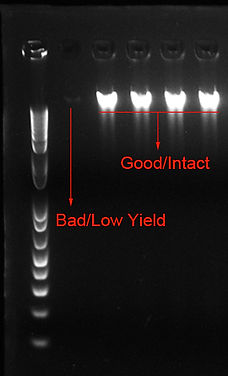
A
Figure 2. Agarose gel image of Intact (A) and Partially Degraded (B) gDNA input.
Note:
-
Samples should appear as a single, intense and sharp band ~23 kb
-
gDNA samples will be classified using the sample classification matrix below
RNA Sequencing Sample Requirements
Illumina MiSeq
-
Submit at least 50 uL of 50 ng/uL Total RNA in water (concentration should be based on QuBit Fluorometer)
-
DNase treatment during RNA isolation is recommended, especially if using a protocol that does not include a polyA purification step.
-
Input RNA must not contain any trace of EDTA, Tween 20, ethanol, or phenol.
-
Samples should have a Bioanalyzer RIN value of >8, an agarose gel profile showing two distinct bands corresponding to the 18S and 28S rRNA, or an MCE-202 MultiNA Trace profile indicative of a high-quality RNA (refer to the figure below).



A
B
C
Figure 3. MCE-202 MultiNA RNA 6000 analysis trace of high-quality RNA (A), partially degraded RNA (B), and fully degraded RNA (C)